Abstract
The ETV6::RUNX1 translocation is the most common chromosomal rearrangement associated with pediatric B cell precursor acute lymphoblastic leukemia (BCP-ALL). As previously shown, ETV6::RUNX1 leads to a distinct clinically silent preleukemic state in utero that manifests in impaired early B cell differentiation. Acquisition of secondary oncogenic mutations gives rise to overt leukemia in 0.2% of children carrying ETV6::RUNX1. In-depth characterization of ETV6::RUNX1+ preleukemia is essential to gain an understanding of leukemic transformation and develop preventive strategies. Here, we identify dysregulation of the epigenetic regulator H1-0 in ETV6::RUNX1+ preleukemia and BCP-ALL.
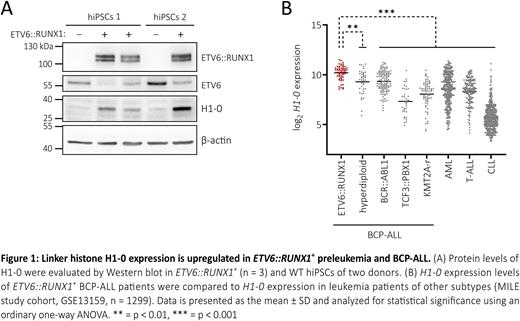
Utilizing CRISPR/Cas9 genome editing, we generated monoclonal ETV6::RUNX1+ human induced pluripotent stem cell (hiPSC) lines with stable ETV6::RUNX1 expression driven by the endogenous ETV6 promoter. To assess the preleukemic setting mediated by ETV6::RUNX1, we performed RNA sequencing (RNA-seq) analysis of ETV6::RUNX1+ and matching wild-type (WT) hiPSCs of two donors. We identified 20 differentially expressed genes in ETV6::RUNX1+ hiPSCs of both donors compared to the respective WT hiPSCs (up: 15, down: 5, absolute fold change ≥ 2, p ≤ 0.05). Interestingly, our RNA-seq analysis revealed consistent upregulation of H1-0 (mean: 2.43-fold), a linker histone involved in chromatin remodeling, cell proliferation and differentiation. Elevated H1-0 levels in the ETV6::RUNX1+ hiPSC lines were confirmed by quantitative real-time PCR and immunoblot (Figure 1A).
To determine H1-0 levels in ETV6::RUNX1+ B cell precursors, we analyzed published gene expression microarray and RNA-seq data sets. Both ETV6::RUNX1+ CD34+ cells (n = 10) derived from umbilical cord blood (Polak et al. Haematologica 2019) and ETV6::RUNX1+ CD34+CD19+ cells (n = 4) generated by hematopoietic differentiation of hiPSCs (Böiers et al. Developmental Cell 2018) showed significant upregulation of H1-0 compared to the respective control cells lacking ETV6::RUNX1 (unpaired t-test, p = 0.0087 and p = 0.0076, respectively). Additionally, we differentiated ETV6::RUNX1+ and WT hiPSCs into CD34+CD45+ hematopoietic progenitor cells and performed single-cell RNA-seq analysis. We identified coexpression of H1-0 with cyclin-dependent kinase inhibitor genes CDKN1A and CDKN1B, indicating a slow or non-cycling phenotype of H1-0+ cells.
Next, we examined H1-0 mRNA levels in gene expression data sets of pediatric and adult leukemia patients (St. Jude PeCan database (n = 876), Jerchel et al. Haematologica 2018 (n = 654), MILE study (n = 1299)). H1-0 expression was found to be significantly elevated in ETV6::RUNX1+ BCP-ALL patient samples compared to other leukemia entities, especially TCF3::PBX1+ ALL (ordinary one-way ANOVA, p ≤ 0.0001), KMT2A rearranged ALL (p ≤ 0.0001) and chronic lymphoblastic leukemia (CLL, p ≤ 0.0001). Strikingly, H1-0 expression was 22.37-fold lower in CLL compared to ETV6::RUNX1+ BCP-ALL (Figure 1B).
This observation prompted us to investigate H1-0 expression in B cell developmental stages using published data sets. Interestingly, we found that H1-0 levels decrease during early B cell development in both normal bone marrow (Black et al. Nucleic Acids Research 2018) and umbilical cord blood-derived cells (Novershtern et al. Cell 2011). Additionally, analysis of single-cell RNA-seq data of early B cell precursors derived from healthy bone marrow (Mehtonen et al. Genome Medicine 2020) and fetal liver (Developmental Cell Atlas Newcastle University) revealed that relative numbers of H1-0 expressing cells per differentiation state decreased along the pseudotime B cell development trajectory.
Taken together, our study identifies H1-0 upregulation as a specific alteration present already at the ETV6::RUNX1+ preleukemic state. High H1-0 expression was associated with slower proliferation and increased stemness. This phenotype might explain the persistence of in utero generated preleukemic ETV6::RUNX1+ clones residing in the bone marrow of approximately 5% of healthy children for years.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.